A Practical Guide to Holo-Omics
About this guidebook
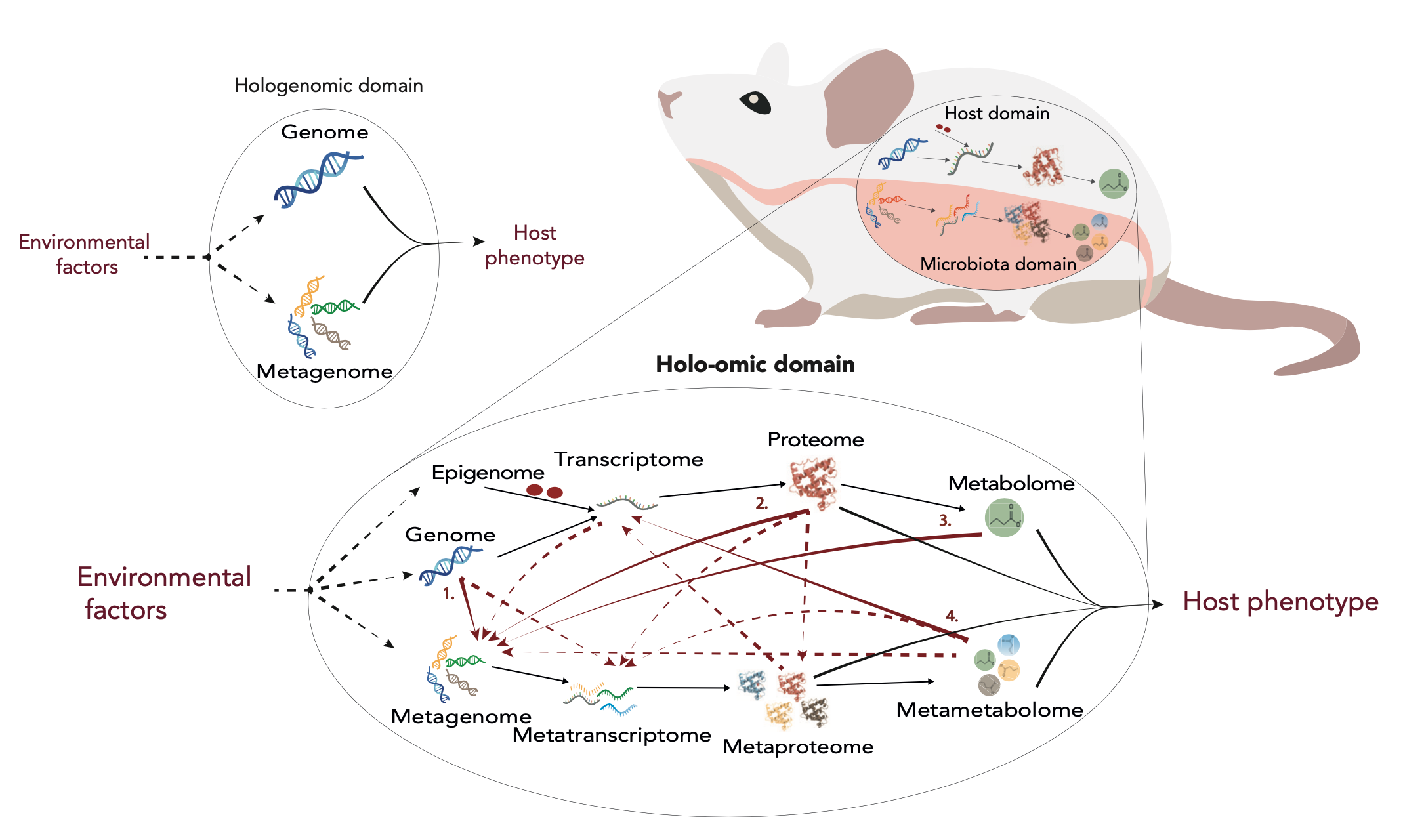
Holo-omics overview. Modified from Nyholm et al. 2020 [1]
The practical guide to holo-omics is a compilation of methodological procedures to generate, analyse and integrate holo-omic data, i.e., multi-omic data jointly generated from hosts and associated microbial communities [1, 2]. This guide extends the contents of the article “A practical introduction to holo-omics”, which aims at guiding researchers to the main critical steps and decision points to perform holo-omic studies. While the article focuses on discussing pros and cons of using multiple available options, the aim of this guide is to compile protocols and pipelines to be implemented by researchers. The practical guide to holo-omics is presented in two formats:
- Website (http://www.holo-omics.science/)
- PDF document (http://www.holo-omics.science/holo_omics_guidebook.pdf)
The guidebook is meant to be a live resource under continuous development, whose contents are updated, replaced and improved as technology and knowledge advances. So, changes are to be expected. However, frozen PDF versions will be released periodically to ensure traceability of the contents.
This guide is presented as a final output of the H2020 project HoloFood. More information about this EU Innovation Action that ran between 2019 and 2023 can be found in the HoloFood Website and the CORDIS website.
Contents
- Introduction: general information about holo-omics, employed data types and study design considerations.
- Laboratory procedures: methods and procedures for generating raw omic data of hosts and microbial communities.
- Bioinformatic procedures: methods and procedures for processing raw omic data into quantitative datasets to be analysed through statistics.
- Statistical procedures: methods and procedures for analysing and integrating holo-omic data.
Protocols, exercises and tutorials
This guide contains example data and bits of code (mostly shell and R) to reproduce data generation and analysis procedures. Code boxes look like the following:
shao4d_perm <- shao4d %>%
tax_transform("identity", rank = "genus") %>%
dist_calc("aitchison") %>%
dist_permanova(
variables = c("breed", "sex", "number_reads"),
n_perms = 99, # you should use more permutations in your real analyses!
n_processes = 1
)
#> Dropping samples with missings: 15
#> 2022-11-24 01:15:20 - Starting PERMANOVA with 99 perms with 1 processes
#> 2022-11-24 01:15:21 - Finished PERMANOVAData sets
The data sets employed in the guidebook are derived from chicken and salmon intestinal samples produced in the H2020 project HoloFood. All the raw omic data, as well as relevant metadata and complementary information can be found in the HoloFood Data Portal.
 This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 817729.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 817729.